Use cases
Use cases objectives:
- Optimize Use Case workloads using machine learning techniques
- Validate the platform in Genomics, Metabolomics, and Surgery use cases with complex pipelines involving data connectors
- Create and validate open libraries of data connectors in the different use cases
Genomics
Creation of methods, fast storage, and communications infrastructures to communicate distributed computing power with scalable storage systems, allowing efficient distribution of datasets across the system.
USE CASE 1. Variants-Interactions Use Case
Complex diseases, such as Type 2 Diabetes (T2D), are caused by the simultaneous effect of multiple genomic variants and other environmental factors. During the last decades, the genomic study of T2D has been broadly approached with diverse methods. However, although hundreds of genomic variants are expected to contribute to disease development, the study of the relation between variants and T2D has been only analyzed in a single independent manner. There are different reasons to avoid this type of analysis. Among them, the analysis of variant interactions and their contribution to developing the disease is an extreme computational problem. Indeed, here we present two use cases where we tackle this type of analysis with different methods:
- Genome-Wide discovery (GWD): use machine learning methods to find groups of variants that, simultaneously, are associated with T2D.
- Multi Dimensionality Reduction (MDR): use statistical methods to discover pairs of variants which, synergically, contribute to the development of T2D.
USE CASE 2. Transcriptomics Use Case
This use case is divided into two main experiments, which can be seen as different use cases given each present different problems and uses different datasets. Such experiments are:
- Transcriptomics Atlas
- Federated Learning for Human Genome Variation Analaysis
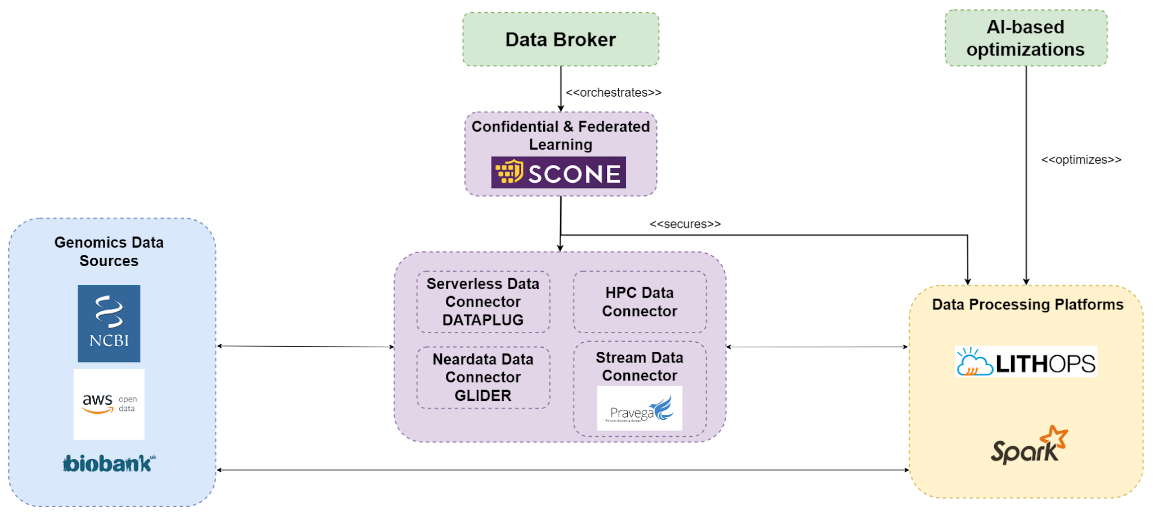
USE CASE 3. Genomics Use Case
This use case is based on variant calling to identify variants from reference genome sequences. We discuss that this type of data is extreme due to the extreme volume of data and extensive pipelines that require. large computational resources to cope with the demands needed to process and analyze this data. The objective of this use case is to port a genomics Variant calling workflow from our HPC implementation to a serverless architecture that allows processing and analyzing extreme genomic data at large scales.
OBJECTIVES:
Variant Calling Pipeline (UKHS):
- Dataplug reduces data transfers by 200%.
- Lithops version is x37.46 faster than HPC version.
Transcriptomics Atlas Use Case (SANO):
- Resource optimization reduces compute cost by 50%.
- Confidential computing with SCONE.
Genomics Epistasis Use Case (BSC):
- MPI version is x5 faster than Apache Spark version.
- HPC Data Connector improves performance by x2.1.
- Resource Auto-scaling with Lithops.
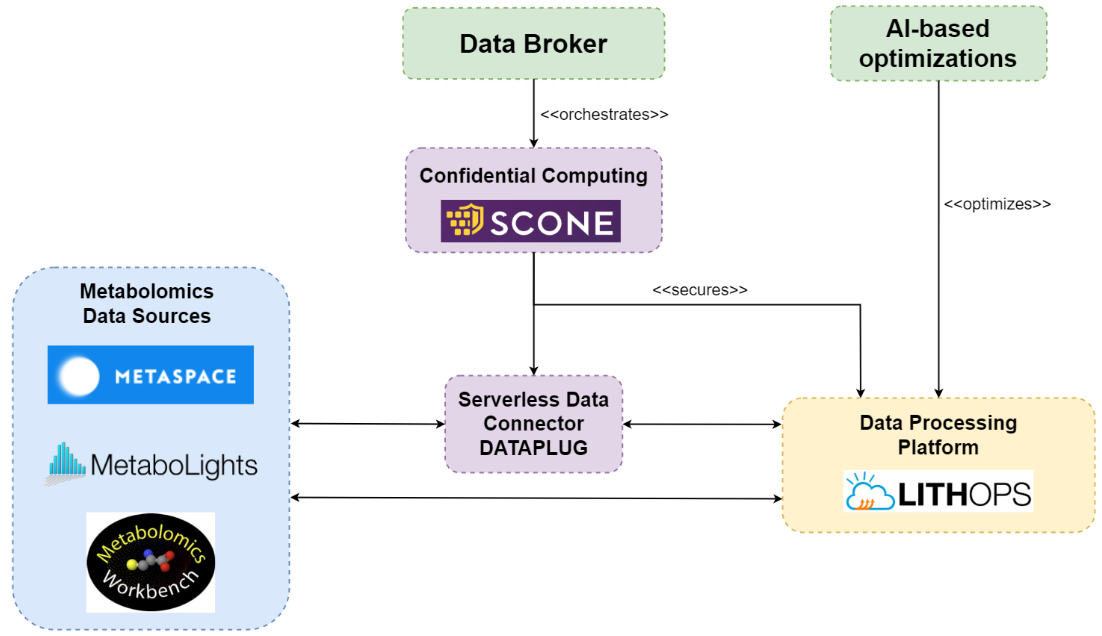
METABOLOMICS
Expand the analysis of metabolomics raw data and boost external access and efficient re-use of open data. Creation of federated and Hybrid distributed architecture and ensuring data privacy but also shared global computations.
USE CASE 1. Metabolomics Use Case
In this use case, we contribute to improving METASPACE, a cloud platform for spatial metabolomics. Spatial metabolomics is a bioanalytical technology for spatially-resolved detection of metabolites, lipids, drugs and other molecules in tissue sections used in biology, medicine, and pharmacology. Spatial metabolomics generates large datasets because for each pixel it produces a mass spectrum containing in the order of 104 dimensions representing abundances of different molecules. A key problem in spatial metabolomics is the metabolite identification or associating the spectral dimensions with specific metabolites they can represent. The individual spatial metabolomics datasets have large sizes, ranging from 1 to 100 GB per tissue section.
In this project, we aim to further improve the open METASPACE cloud platform and strengthen its position as a International Health Data Space.
The overview of the METASPACE cloud platform engine and International Data Space outlining key functionality and types of usage:
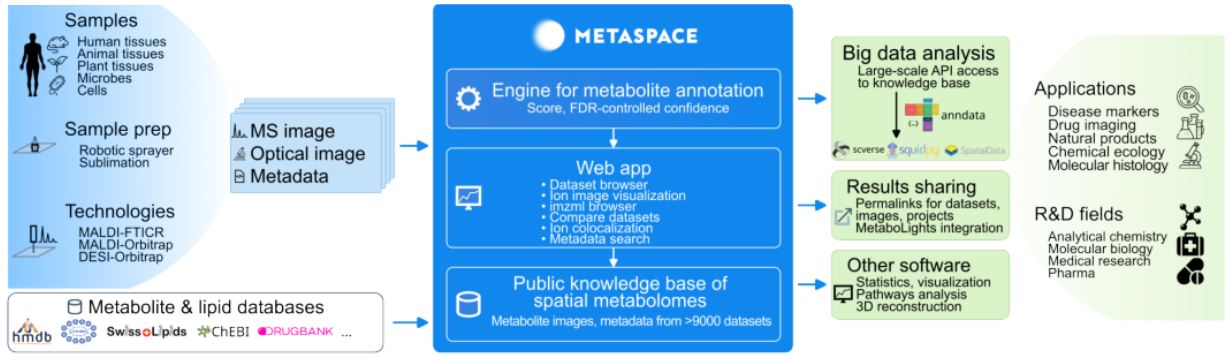
OBJECTIVES. Metabolomics Use Case (EMBL):
- Dataplug offers partitioning strategies for metabolomics data formats (ImzML).
- Resource auto-scaling with Lithops (Datasets from under 1GB to 20GB).
- Confidential computing with SCONE.
- The ML-based metabolite identification is already available to users in the production version of METASPACE and is already used by METASPACE users.
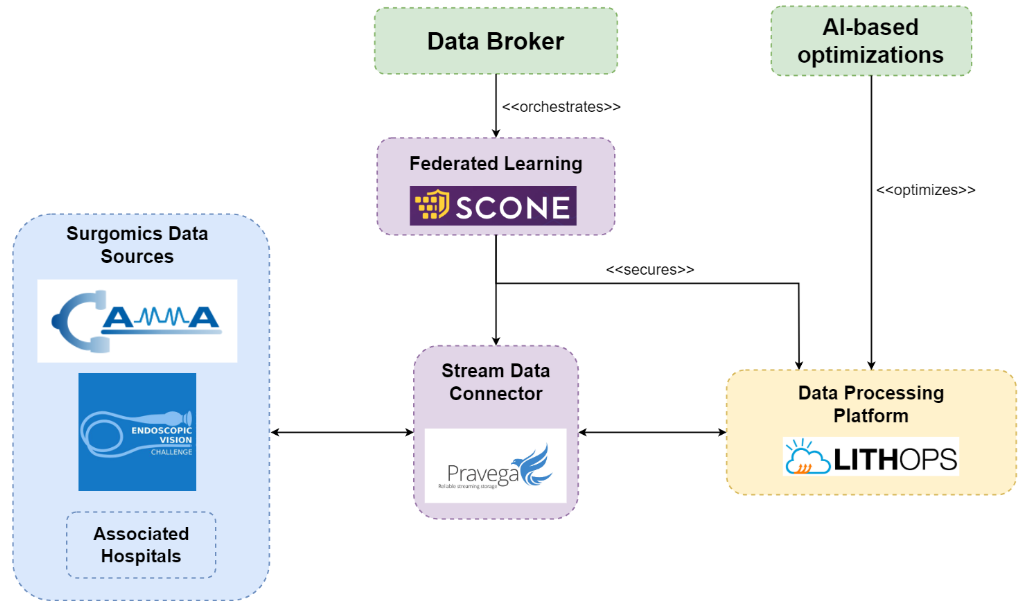
SURGERY
Create generalised machine-learning models that can aid surgeons during surgery and allow video data to be analysed in real-time and with low latency.
USE CASE 1. Surgery Use Case
Our use case in surgery encompasses two distinct focal points. Firstly, we address the challenges inherent in Federated Learning, with a primary objective of improving its security protections. To achieve this, we leverage the Flower framework, enclosing its functionalities within a Docker container for streamlined deployment and management. Additionally, we integrate Scone to ensure security, ensuring the preservation of privacy throughout the Federated Learning processes.
Secondly, we embark on the development of a surgical video streaming application to handle multiple inference jobs. Our strategy revolves around the integration of Pravega and GStreamer into our existing pipelines. This integration empowers us to harness the advanced processing capabilities offered by GStreamer plugins, including our developed segmentation, phase detection, and tool detection plugins. Furthermore, by integrating both GStreamer and Pravega into our infrastructure, we establish a robust ecosystem that contributes to efficient data collection, processing, and storage within the context of surgical video streaming.
OBJECTIVES. Surgery Use Case (NCT):
- Pravega reduces end-to-end IO latency by 45%.
- Confidential computing with SCONE.
- Combined deployment and data management time for video analytics is reduced by 50%.